Kaplan-Meier (KM) plot function for anchored and unanchored cases using ggplot
Source:R/plot_km2.R
kmplot2.RdThis is wrapper function of basic_kmplot2.
The argument setting is similar to maic_anchored and maic_unanchored,
and it is used in those two functions.
Usage
kmplot2(
weights_object,
tte_ipd,
tte_pseudo_ipd,
trt_ipd,
trt_agd,
trt_common = NULL,
normalize_weights = FALSE,
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
km_conf_type = "log-log",
km_layout = c("all", "by_trial", "by_arm"),
time_scale,
...
)Arguments
- weights_object
an object returned by
estimate_weight- tte_ipd
a data frame of individual patient data (IPD) of internal trial, contain at least
"USUBJID","EVENT","TIME"columns and a column indicating treatment assignment- tte_pseudo_ipd
a data frame of pseudo IPD by digitized KM curves of external trial (for time-to-event endpoint), contain at least
"EVENT","TIME"- trt_ipd
a string, name of the interested investigation arm in internal trial
dat_igd(real IPD)- trt_agd
a string, name of the interested investigation arm in external trial
dat_pseudo(pseudo IPD)- trt_common
a string, name of the common comparator in internal and external trial, by default is NULL, indicating unanchored case
- normalize_weights
logical, default is
FALSE. IfTRUE,scaled_weights(normalized weights) inweights_object$datawill be used.- trt_var_ipd
a string, column name in
tte_ipdthat contains the treatment assignment- trt_var_agd
a string, column name in
tte_pseudo_ipdthat contains the treatment assignment- km_conf_type
a string, pass to
conf.typeofsurvfit- km_layout
a string, only applicable for unanchored case (
trt_common = NULL), indicated the desired layout of output KM curve.- time_scale
a string, time unit of median survival time, taking a value of 'years', 'months', weeks' or 'days'
- ...
other arguments in
basic_kmplot2
Value
In unanchored case, a KM plot with risk set table. In anchored case, depending on km_layout,
if "by_trial", 2 by 1 plot, first all KM curves (incl. weighted) in IPD trial, and then KM curves in AgD trial, with risk set table.
if "by_arm", 2 by 1 plot, first KM curves of
trt_agdandtrt_ipd(with and without weights), and then KM curves oftrt_commonin AgD trial and IPD trial (with and without weights). Risk set table is appended.if "all", 2 by 2 plot, all plots in "by_trial" and "by_arm" without risk set table appended.
Examples
# unanchored example using kmplot2
data(weighted_sat)
data(adtte_sat)
data(pseudo_ipd_sat)
kmplot2(
weights_object = weighted_sat,
tte_ipd = adtte_sat,
tte_pseudo_ipd = pseudo_ipd_sat,
trt_ipd = "A",
trt_agd = "B",
trt_common = NULL,
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
time_scale = "month",
break_x_by = 2,
xlim = c(0, 20)
)
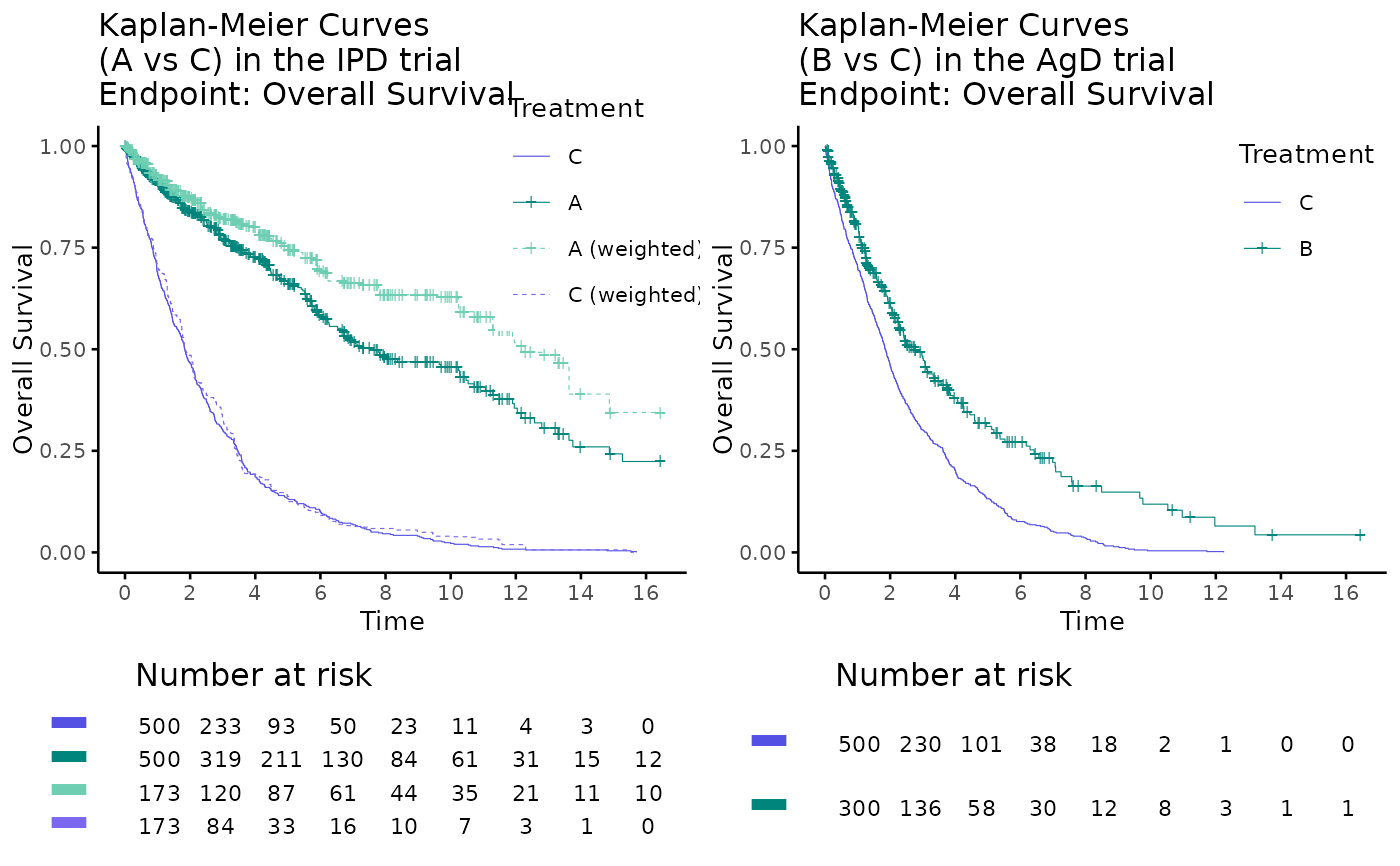
 # anchored example using kmplot2
data(weighted_twt)
data(adtte_twt)
data(pseudo_ipd_twt)
# plot by trial
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "by_trial",
time_scale = "month",
break_x_by = 2
)
# anchored example using kmplot2
data(weighted_twt)
data(adtte_twt)
data(pseudo_ipd_twt)
# plot by trial
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "by_trial",
time_scale = "month",
break_x_by = 2
)
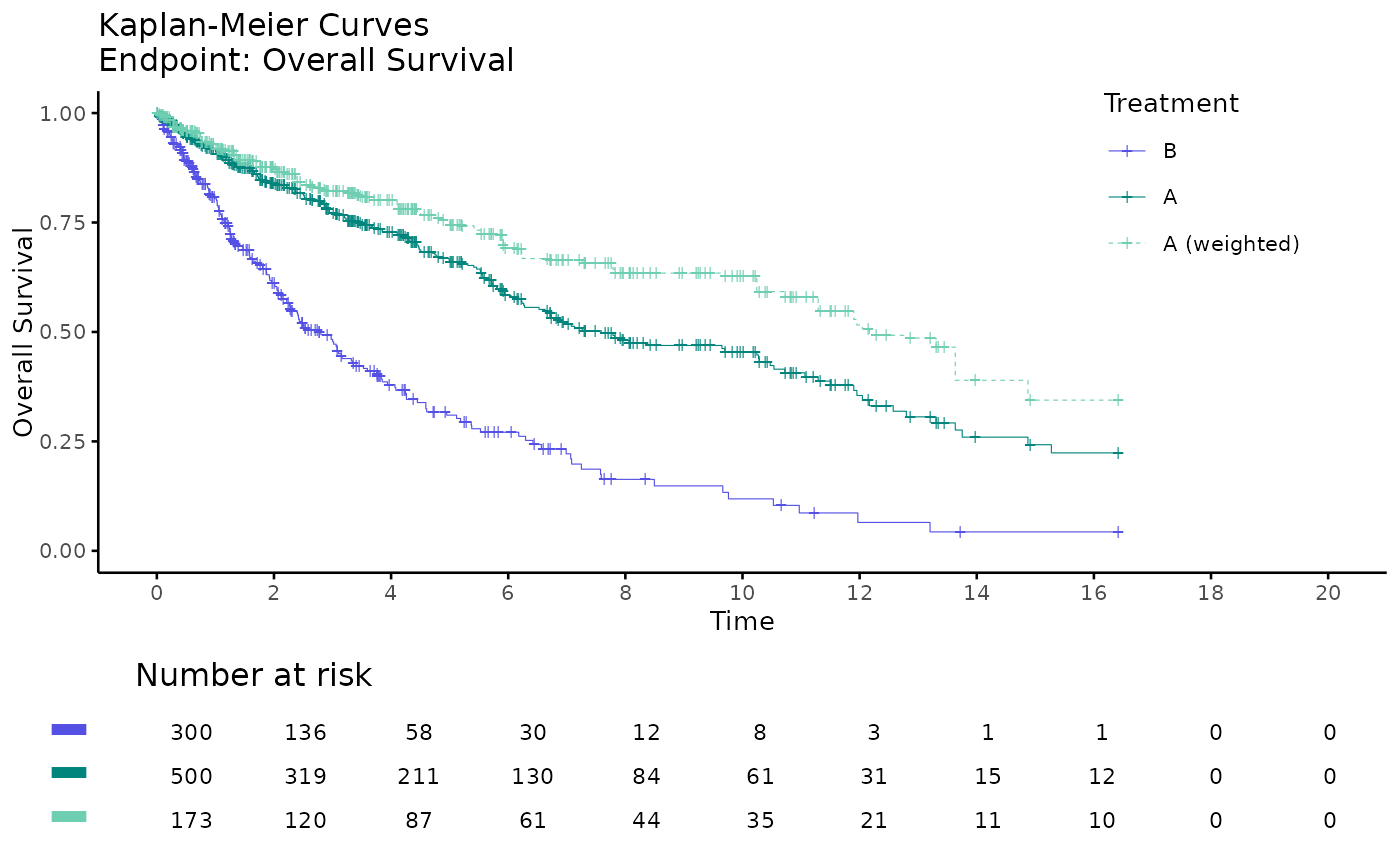
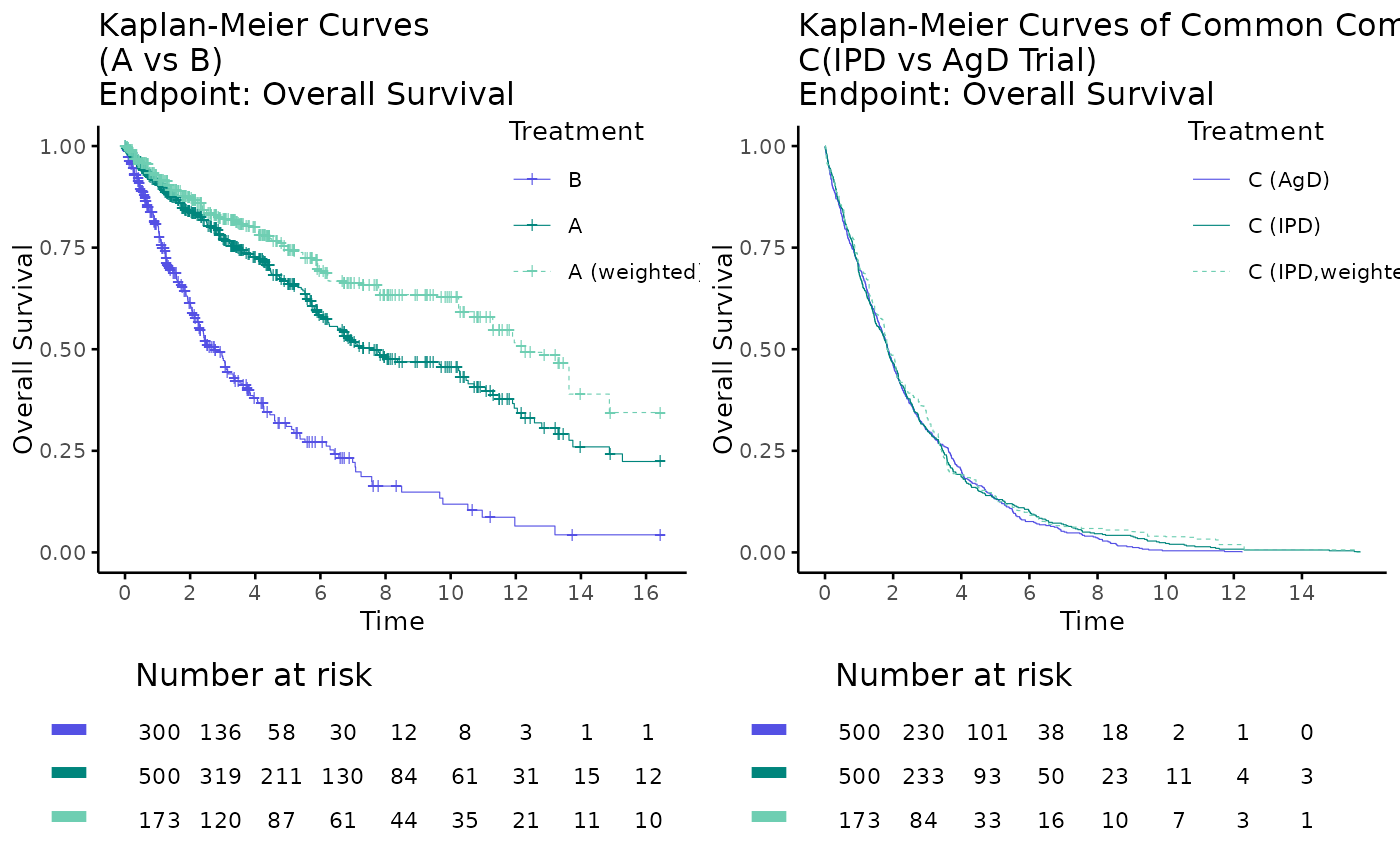
 # plot by arm
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "by_arm",
time_scale = "month",
break_x_by = 2
)
# plot by arm
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "by_arm",
time_scale = "month",
break_x_by = 2
)
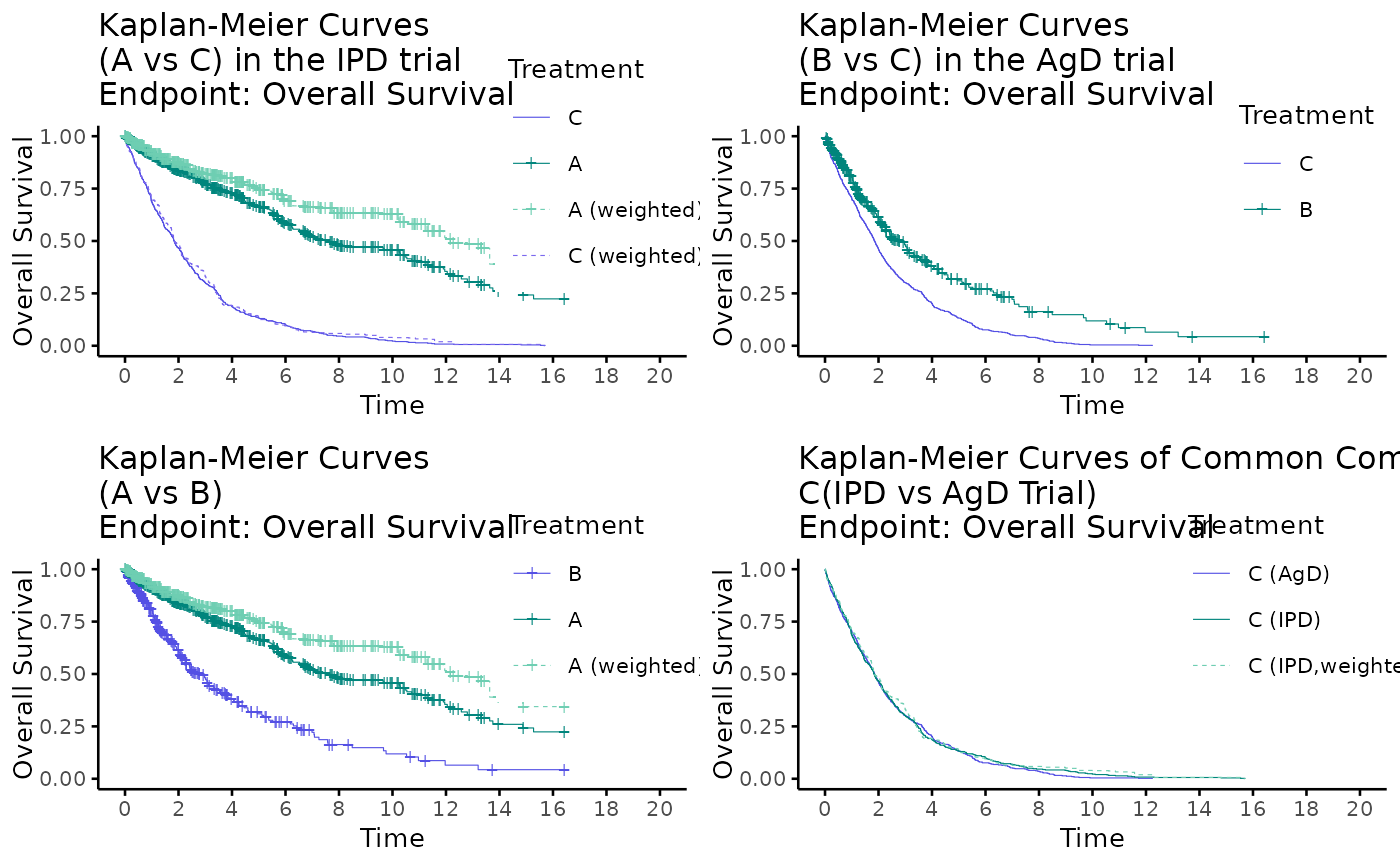
 # plot all
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "all",
time_scale = "month",
break_x_by = 2,
xlim = c(0, 20),
show_risk_set = FALSE
)
# plot all
kmplot2(
weights_object = weighted_twt,
tte_ipd = adtte_twt,
tte_pseudo_ipd = pseudo_ipd_twt,
trt_ipd = "A",
trt_agd = "B",
trt_common = "C",
trt_var_ipd = "ARM",
trt_var_agd = "ARM",
endpoint_name = "Overall Survival",
km_conf_type = "log-log",
km_layout = "all",
time_scale = "month",
break_x_by = 2,
xlim = c(0, 20),
show_risk_set = FALSE
)